DNA analysis can be done by using different methods (1). However, the quality/purity of DNA needed for such analysis depends on type of analysis we are doing. We can analyze both human DNA and bacterial DNA by using methods like PCR, restriction fragment length polymorphism (RFLP) or sequencing. So, the most critical part is the purification of DNA for these kind of analysis (2). We can purify DNA from the reagents generally present in our lab. However, we will send our purified DNA for sequencing. So, we need highly purified DNA (3).
The common procedure for purification of DNA is the disruption of cells creating a lysate, separating the DNA from cellular debris including proteins and RNA. The traditional home-made preps uses phenol: chloroform method followed by ethanol precipitation of DNA. Phenol is organic non-polar compound. So, it helps in separation of aqueous phase and organic phase in the solution. As the DNA is highly polar, it does not dissolve in phenol. Also, chloroform is used to make the solution further denser as both phenol (1.07 g/cm3) and chloroform (1.47 g/cm3) are denser than water and chloroform is easily mixed with phenol. This mixture also precipitates the protein better than either of the compound alone. DNA and RNA are separated by exploiting the difference in pH (4) of phenol. However, DNA separated by home-made preps might have some contamination which is not desirable in sequencing. So, we are using Wizard Plus SV Miniprep kit from the Promega Corporatin to purify our plasmid from the host cell.
The plasmid purification from Promega’s kit is based on silica. The host cell pellet is obtained by centrifugation and the pellet is re-suspended in cell resuspension solution. This solution contains chaotropic salts or detergents which denature the host cells (2). Sodium dodecyl sulfate (SDS) and NaOH are mostly used. SDS solubilize the cell membrane and NaOH break down the cell wall of the host cells. This NaoH also converts the circular plasmid and genomic DNA in to single stranded DNA. After that alkaline protease is added to this mixture to denature the cellular proteins. Further, this alkaline solution is neutralized by adding neutralization solution (NSB). This NSB contains potassium acetate which reduces the alkalinity of the solution mixture. Under these condition, the single stranded DNA (plasmid) re-natured to double stranded DNA. Genomic DNA is not re-natured due to its large size. However, if the genomic DNA is broken into smaller fragments, it could re-nature itself and contaminate our plasmid. The double-stranded plasmids remain dissolved in solution mixture while the single stranded genomic DNA, the denatured protein and the SDA aggregate together by hydrophobic interactions and form white precipitate. The plasmid is separated from this precipitate by centrifugation (3, 5). Thus obtained plasmid is further cleaned up by washing with column wash solution (CWA) and centrifugation. Finally, the plasmid DNA is eluted by using nuclease free water (2, 3) and purified plasmid is obtained.
The concentration of thus purified DNA is measured by spectrophotometer at 260 nm – 280 nm.
Restriction digestion and gel analysis is done to confirm the isolated DNA and the insert of our interest (3). To analyze the cloned DNA, we will digest the plasmid with restriction enzymes. Restriction enzymes are endonucleases found in bacteria that cut a DNA molecule at a specific site. These cut could be sticky end or blunt end. These enzymes are part of defense mechanism in these organisms which is used against foreign DNA like phage DNA (3, 6). Restriction enzymes have wide applications in molecular biology. These can be used for DNA mapping, making recombinant DNA molecules, cloning, and identification of individuals or specific strain of bacteria (3, 7).
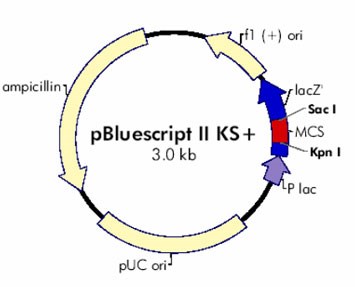
Our sequence of interest is inserted between EcoR I site and Xho I site. So, we will use EcoR I and Xho I restriction enzymes to cut out our sequence. EcoR I is obtained from E. coli RY13. ‘R’ denotes the strain of bacteria while letter ‘I’ indicate that it was the first of its type extracted from the organism. It cuts at 5’…..G^AATTC…..3’ palindromic site of DNA (8, 9, 10). Xho I restriction enzyme is obtained from E. coli bacteria that carries Xho I gene of Xanthomonas holcicola (11). It digest the DNA molecules at 5’… C^TCGAG…3’ palindromic site (12).
After digestion of the plasmid with EcoRI and XhoI, we will run the digested fragments of plasmid on agarose gel to determine the sizes of our digestion product. We will use BlueJuice as the tracking dye because now we know that our insert sequence is greater than 300 bp.
References:
1. Hinkley E (2017). What is DNA analysis? https://dnatestingchoice.com/en-us/news/2017-03-21-what-is-dna-analysis (accessed on Sept 17, 2017).
2. Promega Corporation (assessed on Sept 17, 2017). DNA purification. https://www.promega.com/resources/product-guides-and-selectors/protocols-and-applications-guide/dna-purification/
3. Dietrich M (2017). Plasmid minipreps for use as sequencing templates. Short Protocols in Molecular Biology. Grand Valley State University. 23-24.
4. Zumbo P (accessed on Sept 17, 2017). Phenol-chloroform extraction. Weill Cornell Medical College. http://physiology.med.cornell.edu/faculty/mason/lab/zumbo/files/PHENOL-CHLOROFORM.pdf
5. Oswald N (accessed Sept 17, 2017). The Basics: How Alkaline Lysis Works. http://bitesizebio.com/180/the-basics-how-alkaline-lysis-works/
6. Arizona State University (accessed on Sept 17, 2017). Restriction Enzymes. https://askabiologist.asu.edu/restriction-enzymes
7. Guilfole P G (accessed on Sept 17, 2017). Restriction Enzymes- Use of Restriction Enzymes in Biotechnology. http://medicine.jrank.org/pages/2779/Restriction-Enzymes-Use-Restriction-Enzymes-in-Biotechnology.html
8. Wikipedia (accessed on Sept 17, 2017). EcoR I. https://en.wikipedia.org/wiki/EcoRI
9. ThermoFisher Scientific (accessed on Sept 17, 2017). AnzaTM11 EcoRI. https://www.thermofisher.com/order/catalog/product/IVGN0116?gclid=CjwKCAjw3f3NBRBPEiwAiiHxGKUmdDOpst8j0PdWsmM5LSzSTCGjgKBBEo12tCuhIuXk1V6od-yayRoC1s8QAvD_BwE&s_kwcid=AL!3652!3!163711299264!b!!g!!%2Becori&mkwid=sy2q4lW0m-dc_pcrid_163711299264_pkw_+ecori_pmt_b_slid__dimid=&ef_id=V2ycSAAAAAYBPed2:20170918150412:s
10. Takara (accessed on Sept 17, 2017). EcoRI. http://www.clontech.com/SN/Products/Molecular_Biology_Tools/Restriction_Enzymes/Restriction_Enzymes_e_f/EcoRI
11. New Englab Biolabs (accessed on Sept 17, 2017). XhoI. https://www.neb.com/products/r0146-xhoi#pd-properties-usage
12. ThermoFisher Scientific (accessed on Sept 17, 2017). FastDigest XhoI. https://www.thermofisher.com/order/catalog/product/FD0694
